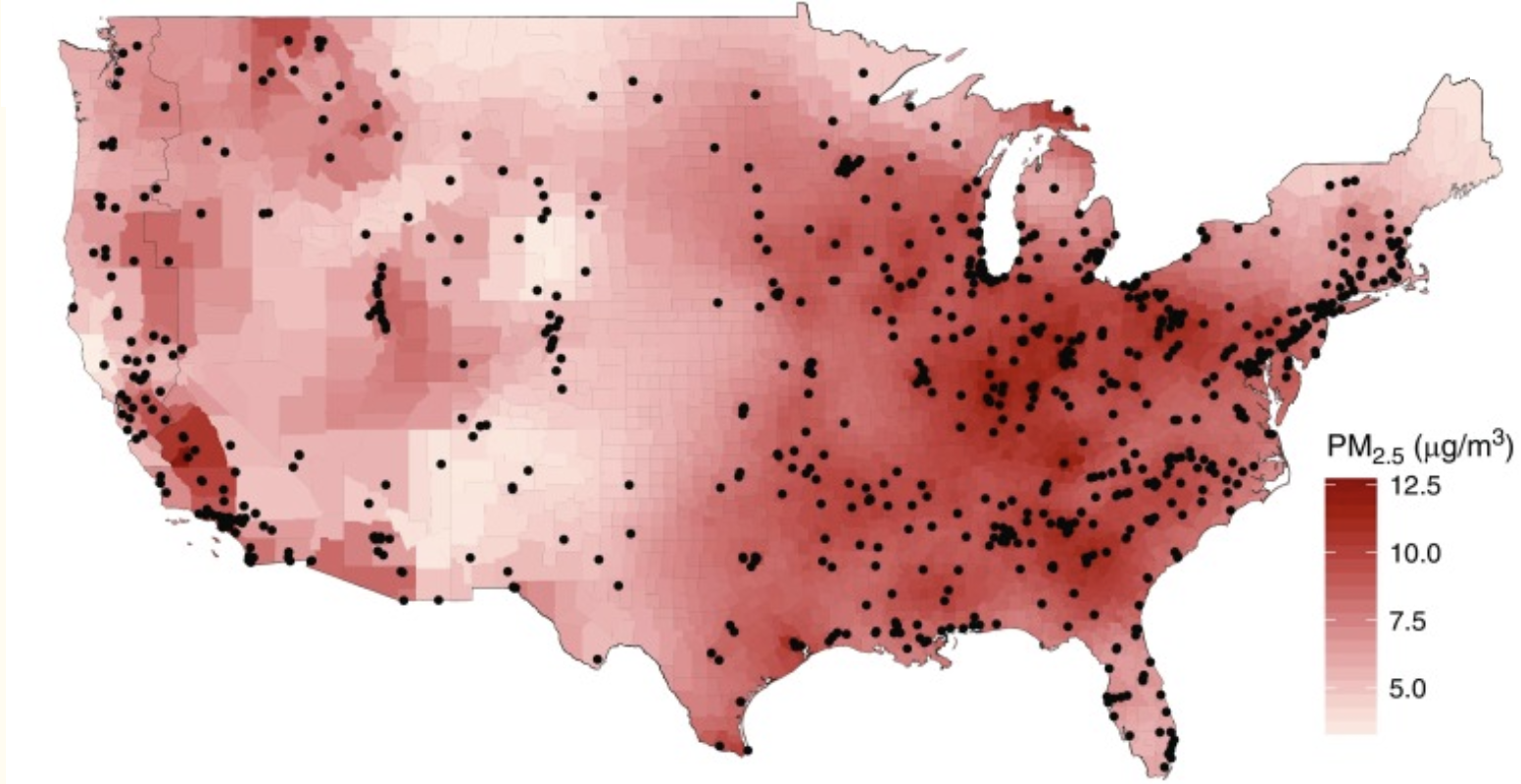
 {width=800 px}
{width=800 px}
Biomedical Open Case Studies: {Title}
Disclaimer: The purpose of the Open Case Studies project is to demonstrate the use of various data science methods, tools, and software in the context of messy, real-world data. A given case study does not cover all aspects of the research process, is not claiming to be the most appropriate way to analyze a given data set, and should not be used in the context of making policy decisions without external consultation from scientific experts. In addition, due to size constraints, datasets used within a case study may be subset of the original/full dataset.
This work is licensed under the Creative Commons Attribution-NonCommerical 3.0 (CC BY-NC 3.0) United States License.
To access the GitHub repository for this case study see here: {LINK}
To cite this case study, please use:
{Fill In Authors}. {(YEAR)}. {github source url}. {Title} {(Version)}.
Motivation
This case study explores {}…
Main Question
Our main question:
- Question 1
- Question 2
Learning Objectives
In this case study, we will explore {}.
This case study will particularly focus on {}.
The skills, methods, and concepts that students will be familiar with by the end of this case study are:
Data Science/Bioinformatics Learning Objectives:
- {FILL IN}
- {FILL IN}
- {FILL IN}
Biological/Topical Learning Objectives:
- {FILL IN}
- {FILL IN}
- {FILL IN}
We will begin by loading the packages that we will need:
#add library() calls
library(here)here() starts at /home| Package | Use |
|---|---|
| {Package Name} | {Package use} |
| {Package Name} | {Package use} |
The first time we use a function, we will use the :: to indicate which package we are using. Unless we have overlapping function names, this is not necessary, but we will include it here to be informative about where the functions we will use come from.
Context
{FILL IN}
What are the data?
| Variable | Details |
|---|---|
| variable1 | Variable info – more details – more detials Example: Content content |
| variable2 | Variable info – more details – more detials Example: Content content |
Limitations
There are some important considerations regarding this data analysis to keep in mind:
- {FILL IN}
- {FILL IN}
Ethical Considerations
There are some important ethical considerations when working with data relating to this case study’s main questions.
- {FILL IN}
- {FILL IN}
Data Import
pm <-readr::read_csv(here("data", "raw", "pm25_data.csv"))Rows: 876 Columns: 50
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (3): state, county, city
dbl (47): id, value, fips, lat, lon, CMAQ, zcta, zcta_area, zcta_pop, imp_a5...
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.save(pm, file = here::here("data", "imported", "pm25_data_imported.rda"))Data Wrangling
If you have been following along but stopped, we could load our imported data like so:
load(here::here("data", "imported", "pm25_data_imported.rda"))If you skipped the data import section click here.
An RDA version (stands for R data) of the data can be found here or slightly more directly here. Download this file and then place it in your current working directory within a subdirectory called “imported” within a directory called “data” to use the following code. We used an RStudio project and the here package to navigate to the file more easily.
load(here::here("data", "imported", "co2_data_imported.rda"))To allow users to skip import and wrangling we will save the data as an RDA file as well as a CSV file as this is often useful to send our data to collaborators. We will save this in a “wrangled” subdirectory of our “data” directory of our working directory.
save(pm, file = here::here("data", "wrangled", "wrangled_data.rda"))
readr::write_csv(pm, file = here::here("data","wrangled", "wrangled_data.csv"))Data Visualization
If you have been following along but stopped, we could load our wrangled data like so:
load(here::here("data", "wrangled", "wrangled_data.rda"))If you skipped the data import section click here.
An RDA file (stands for R data) of the data can be found here or slightly more directly here. Download this file and then place it in your current working directory within a subdirectory called “wrangled” within a subdirectory called “data” to use the following code. We used an RStudio project and the here package to navigate to the file more easily.
load(here::here("data", "wrangled", "wrangled_data.rda"))Data Analysis
If you have been following along but stopped, we could load our wrangled data like so:
load(here::here("data", "wrangled", "wrangled_data.rda"))If you skipped the data import section click here.
An RDA file (stands for R data) of the data can be found here or slightly more directly here. Download this file and then place it in your current working directory within a subdirectory called “wrangled” within a subdirectory called “data” to use the following code. We used an RStudio project and the here package to navigate to the file more easily.
load(here::here("data", "wrangled", "wrangled_data.rda"))Summary
Synopsis
Summary Plot
Suggested Homework
Additional Information
Helpful Links
Terms and concepts covered:
Packages used in this case study:
Session Info
devtools::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.3.2 (2023-10-31)
os Ubuntu 22.04.4 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate en_US.UTF-8
ctype en_US.UTF-8
tz Etc/UTC
date 2025-10-21
pandoc 3.1.1 @ /usr/local/bin/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
bit 4.0.5 2022-11-15 [1] RSPM (R 4.3.0)
bit64 4.0.5 2020-08-30 [1] RSPM (R 4.3.0)
cachem 1.0.8 2023-05-01 [1] RSPM (R 4.3.0)
cli 3.6.2 2023-12-11 [1] RSPM (R 4.3.0)
crayon 1.5.2 2022-09-29 [1] RSPM (R 4.3.0)
devtools 2.4.5 2022-10-11 [1] RSPM (R 4.3.0)
digest 0.6.34 2024-01-11 [1] RSPM (R 4.3.0)
ellipsis 0.3.2 2021-04-29 [1] RSPM (R 4.3.0)
evaluate 1.0.4 2025-06-18 [1] CRAN (R 4.3.2)
fansi 1.0.6 2023-12-08 [1] RSPM (R 4.3.0)
fastmap 1.1.1 2023-02-24 [1] RSPM (R 4.3.0)
fs 1.6.3 2023-07-20 [1] RSPM (R 4.3.0)
glue 1.7.0 2024-01-09 [1] RSPM (R 4.3.0)
here * 1.0.1 2020-12-13 [1] CRAN (R 4.3.2)
hms 1.1.3 2023-03-21 [1] RSPM (R 4.3.0)
htmltools 0.5.7 2023-11-03 [1] RSPM (R 4.3.0)
htmlwidgets 1.6.4 2023-12-06 [1] RSPM (R 4.3.0)
httpuv 1.6.14 2024-01-26 [1] RSPM (R 4.3.0)
jsonlite 1.8.8 2023-12-04 [1] RSPM (R 4.3.0)
knitr 1.50 2025-03-16 [1] CRAN (R 4.3.2)
later 1.3.2 2023-12-06 [1] RSPM (R 4.3.0)
lifecycle 1.0.4 2023-11-07 [1] RSPM (R 4.3.0)
magrittr 2.0.3 2022-03-30 [1] RSPM (R 4.3.0)
memoise 2.0.1 2021-11-26 [1] RSPM (R 4.3.0)
mime 0.12 2021-09-28 [1] RSPM (R 4.3.0)
miniUI 0.1.1.1 2018-05-18 [1] RSPM (R 4.3.0)
pillar 1.9.0 2023-03-22 [1] RSPM (R 4.3.0)
pkgbuild 1.4.3 2023-12-10 [1] RSPM (R 4.3.0)
pkgconfig 2.0.3 2019-09-22 [1] RSPM (R 4.3.0)
pkgload 1.3.4 2024-01-16 [1] RSPM (R 4.3.0)
profvis 0.3.8 2023-05-02 [1] RSPM (R 4.3.0)
promises 1.2.1 2023-08-10 [1] RSPM (R 4.3.0)
purrr 1.0.2 2023-08-10 [1] RSPM (R 4.3.0)
R6 2.5.1 2021-08-19 [1] RSPM (R 4.3.0)
Rcpp 1.0.12 2024-01-09 [1] RSPM (R 4.3.0)
readr 2.1.5 2024-01-10 [1] RSPM (R 4.3.0)
remotes 2.4.2.1 2023-07-18 [1] RSPM (R 4.3.0)
rlang 1.1.6 2025-04-11 [1] CRAN (R 4.3.2)
rmarkdown 2.25 2023-09-18 [1] RSPM (R 4.3.0)
rprojroot 2.1.0 2025-07-12 [1] CRAN (R 4.3.2)
sessioninfo 1.2.2 2021-12-06 [1] RSPM (R 4.3.0)
shiny 1.8.0 2023-11-17 [1] RSPM (R 4.3.0)
stringi 1.8.3 2023-12-11 [1] RSPM (R 4.3.0)
stringr 1.5.1 2023-11-14 [1] RSPM (R 4.3.0)
tibble 3.3.0 2025-06-08 [1] CRAN (R 4.3.2)
tidyselect 1.2.0 2022-10-10 [1] RSPM (R 4.3.0)
tzdb 0.4.0 2023-05-12 [1] RSPM (R 4.3.0)
urlchecker 1.0.1 2021-11-30 [1] RSPM (R 4.3.0)
usethis 2.2.3 2024-02-19 [1] RSPM (R 4.3.0)
utf8 1.2.4 2023-10-22 [1] RSPM (R 4.3.0)
vctrs 0.6.5 2023-12-01 [1] RSPM (R 4.3.0)
vroom 1.6.5 2023-12-05 [1] RSPM (R 4.3.0)
xfun 0.52 2025-04-02 [1] CRAN (R 4.3.2)
xtable 1.8-4 2019-04-21 [1] RSPM (R 4.3.0)
yaml 2.3.10 2024-07-26 [1] CRAN (R 4.3.2)
[1] /usr/local/lib/R/site-library
[2] /usr/local/lib/R/library
──────────────────────────────────────────────────────────────────────────────